Hundreds of millions of dollars have been spent on comparing the frequencies of genetic variants in disease-afflicted and control populations to suss out the genetic basis of diseases, common or rare. The results for diseases with diabetes have been mixed which despite the high concordance of this disease in identical twins raises yet again the question of the role of the environment in the etiology of diabetes. Of course, we know that the inherited component of diabetes risk is contingent on environmental factors (notably diet) but these are much harder to quantify and moreover there is a universe of environmental risks that is potentially much larger than the entirety of the genome. So how to go about capturing more of the environmental risk as it pertains to real human beings (as opposed to petri dish or rodent studies)?
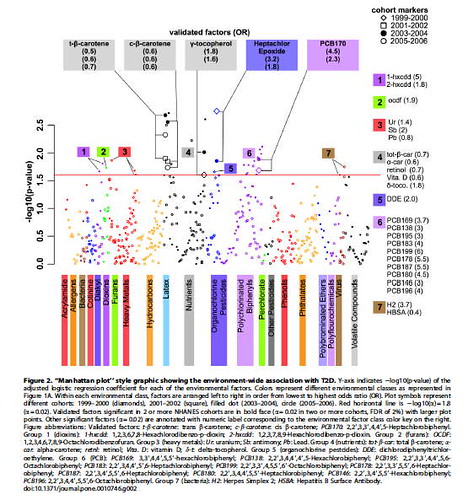
Atul Butte at Stanford shows us how, through the lens of informatics, systematic approaches can be taken to understand the environmentally-borne determinants of our disease burden. He leverages one of the most admirable public national studies of US citizens, the NHANES study. Butte and colleagues performed analyses analoguously to genome wide association studies but instead examined each of the chemicals (e.g. pesticides, heavy metals, vitamin metabolites) measured in the urine and blood of the members of the NHANES groups comparing cases of diabetes vs. controls. As summarized in the figure below, after comparing multiple groups they found a consistent increased risk of diabetes with PCB and pesticide derivatives and a decreased risk with some carotene derivatives (related to vitamin A). Although, just as for their genome-wide analogues, the results of this Environment Wide Association Study (EWAS) should be taken with due caution, they add to a growing array of methodologies that provide a complement to conventional randomized and controlled studies.
No comments:
Post a Comment