A very informative interview with our very own Ben Adida by Yahoo! regarding RDFa. This is a format that will allow very lightweight incorporation of structured data within web pages that will allow the kinds of interoperable applications we are developing at the Harvard Catalyst far easier to build and disseminate.
Harvard Medical School's Countway Library of Medicine and the new frontiers in biomedical computing
2008-12-20
2008-12-18
Can we afford NOT to invest in basic research?
With thanks to Ted Shortliffe who pointed out this article by Bill Buxton. Bill makes an interesting argument in favor of basic research which in many ways runs counter to trends in federal and corporate funding.
2008-12-15
Foundations of Policy
"Twenty-first century leaders in medicine and government are confronted by questions of enormous magnitude: What are the determinants of disease and its distribution? How should health outcomes be measured? How are we to optimize health care delivery and financing, and how are we to ensure access to the fruits of medical science to the poor of this country and the developing world? However, such twenty-first century dilemmas are not new."
The Center of the History of Medicine of the Countway Library has been growing under the leadership of Scott Podolsky and Kathryn Baker Hammond and most recently they were awarded a grant from the Andrew Mellon Foundation that will enable, for the first time, research in the manuscript collections of four influential leaders in public health: Leona Baumgartner, Alan Macy Butler, Howard Hiatt, and David Rutstein. This adds to the growing list of new initiatives by the Center, An important step towards understanding the current and future challenges in public health.
Can we understand genetics? Can you help?
In the context of the multitudinous business plans of various direct-to-consumer genomics companies, any informed analyst should wonder: Can we (healthcare consumers) understand the information communicated by the long-promised, now available, large personalized genomic data sets? This in the context of much evidence that doctors cannot correctly interpret genetic tests and can be readily influenced by genetic testing companies.
With funding from the NIH, we are trying to study how to have patients manage, control and understand their own genomic data. There are several ways you can help but perhaps most importantly it is those who have had experience as consumers of genetics counseling who could help by serving as study subjects to determine what works and what does not in web-borne computer interfaces for direct-to-consumer disclosure of genetic risk data.
2008-12-05
Crossing the chasm from in silico to in vivo
Atul Butte just published a nice paper which illustrates how biomedical informaticians can mine large public (gene expression) data bases to identify novel gene variants with some relevance to human disease. He then goes on to outsource his biological validation to a collaborator half way around our planet to show in vivo that these database driven leads can be reproduced independently. As many aspects of biological discovery become commoditized, it's work like this that reminds us that it is those who as the right questions and who can reach out for the right team to answer them. It should be an important goal of translational science to support agility in this kind of multidisciplinary science-swarming.
2008-11-26
iFail
Even in the rarefied reaches of the ivory tower, you will find various previously productive scientists with their hands under the table banging on their personal communication devices. In that context, this report on the failure rate of these devices may be of interest. Nonetheless, there does appear to be real functionality that is well suited for scholarly communication and education.
2008-11-24
Primary care burdened by primary data
This report out of the Physicians' Foundation shows that doctors are dissatisfied, are going to quit in droves and 94% said the time they devote to non-clinical paperwork in the last three years has increased. So why does it seem that so many are still trying to get into medical school? Is there a medical practice reality chasm?
'Spinning' up the tissues at Harvard
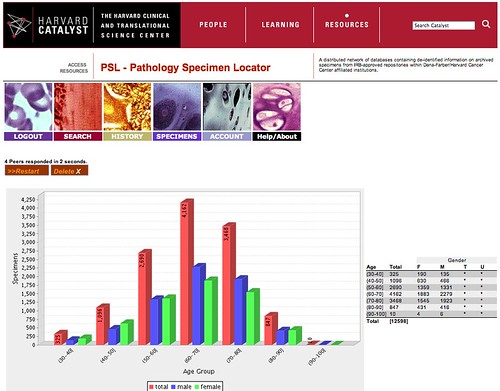
Ever since the late 1990's we have been working on a variety of methods for retrieving data across disparate institutions that are often not even part of the same corporation. In response to an RFA from the National Cancer Institute, called the Shared Pathology Informatics Network, we developed a toolkit that is now in widespread use, specifically to enable genomic and other biological studies on the millions of specimens that are archived across healthcare institutions. As is often the case, we were late in bringing our own tools to use in our own backyard, but that has now happened. The Pathology Specimen Locator (PSL) is now live at our CTSA Catalyst portal. As shown in this screenshot, with authorized credentials, I was able to see that there are over 10,000 lung cancer samples (you can query for any tissue type and disease) across a wide range of ages. It is this sort of IRB-protected, informatics-enabled data liquidity that will accelerate our translational research efforts. Hats off to the entire team but particularly Andy, Frank, John, and Mark.
Transcriptome of a Trio
Hugh Rienhoff will be speaking about an interesting intersection of expression and genetic transmission studies on December 4th.
2008-11-16
Thousands of free books
As per the Lexcyle website "Stanza is a free application for your iPhone and iPod Touch. Use it to download from a vast selection of over 40,000 books and periodicals, and read them right on your phone. It’s a wireless electronic library that stays open 24/7." A quick search for science fiction reveals classics by H. G. Wells and pulp fiction by "Doc" E. E. Smith. All without Digital Rights Management. A real treat.
Images in Development
If an image is worth a thousand words, and a video is 30 frames per second, then these movies of a developing zebrafish must be worth millions. I could not conceive now of a course in embryogenesis that did not include these fascinating and wholistic perspectives on the developing organism. What drives those pulses of development, those three- to four-cell cabals, those intricate dances of the somites? Is it all gradients and cell-cell interactions? Or is there a master control program?
2008-11-12
Matching high-throughput genomics to high-throughput mining of the literature
In this study (alas, for-fee-access) , Dennis Wall demonstrates how to mine the literature (aka the biomedical bibliome) to focus the analyses of noisy genomic modalities which by virtue of measuring thousands of genes have to be aggressively corrected for multiple hypothesis testing [ed. Disclosure: I am a co-author]. By examining gene expression analyses of individuals with autism through the lens of the prior literature on neuro-psychiatric-behavioral disorders, he is able to identify genes significantly differentially expressed in individuals with autism, both known and previously non-implicated genes. This is one of a growing list of publications that are attempting to match the high throughput qualities of genomic measurements with an equally efficient automated "reading" of all the painstakingly obtained biomedical investigational literature. It also suggests that an even more detailed annotation by librarians of the existing literature (analogously to what the National Library of Medicine has done for years for the broad addition of meta data) will be productively leveraged in future investigations. I suppose this is where my colleagues from the Semantic Web have another opportunity to feed the search engines of Google.
2008-11-06
When consent gets in the way
"When consent gets in the way" is the purposefully provocative title of the article (alas, freely open to the public for only 1 month) by Patrick Taylor in which he questions the current dogma about the relationship between consent, privacy, ethical behavior and the public good. Although I was among the early promoters of a strong model of patient personal control of their healthcare data and healthcare decision-making, I found several of Taylor's points compelling. Notably, that "it is questionable whether consent-for-everything will promote privacy and public trust" and "There is more to ethical decision-making than asking whether decisions are made autonomously. Do they take into account virtues, moral values and human narratives with less impoverished conceptions of human freedom? Are the choices good, and do they respect ethical obligations to others?" These points are at the heart of current trends in increasing the "liquidity of patient data" and are certainly central to the business plans of several large companies. How we respond to these assertions as a individuals and a society will be telling. Not only for our research policies and infrastructure but for our conception of healthcare.
2008-11-04
E-science = Librarians 2.0?
In this article (free, but requires registration), in The Scientist, we are given another glimpse into the human components of E-science. I'ts one thing to have a distributed computational network capable of delivering teraflops at the tap of a button, it's another thing to have a biological scientific workforce that knows how to use those cycles at all, let alone effectively. Librarians such as our very own David Osterbur, who are also trained in the biological sciences (and particularly in the use of bioinformatics tools) are a remarkably efficient means to help disseminate a working knowledge of these essential tools to our local communities of biological investigators and beyond.
2008-10-30
The Joy of Collecting
Every once in a while, I get a notice that reminds me that there are pleasant avocations that don't quite make it to "Reality TV" fare. Here is one such announcement.
2008-10-28
Google wins?
Google just settled a lawsuit brought against them by authors and publishers. At first blush, it seems that Google just agreed to spend $125 million to avoid even greater financial liability. But, as Ben Reis pointed out to me, they only had to invest $125M to get the book publishing equivalent of the iTunes store agreed to by a large swathe of publishers. Interesting jujutsu.
2008-10-27
Proceeding with precedings
An interesting subfractionation of the open access space is the Nature Precedings. No peer review but broad visibility and the ability to drive a stake in a scientific claim in a very clear way. In many ways this is as revisiting of the Physics preprint service that was among the original drivers of the architecting of the Web.
Plagiarism-proof
With all the concern about plagiarism, it is refreshing to read this essay on the cultivation of creativity and intellectual self-reliance that masquerades as a recipe for plagiarism prevention. Not that it does not outline some ingenious heuristics to prevent plagiarism. It does, by suggesting the assignment of topics that are uniquely at the intersection of the individual's experience and local identity, so as to defy any generality that would allow easy textual cloning to substitute for reflection and crafted writing. It's quite impressive to witness, even if from afar, the dedication to shaping personalized educational experiences that are broadly informed. With such teachers plagiarism really does seem beside the point.
Finally, and most satisfying, is that this essay is a chapter in a fully open access book that is part of an open access series from University of Michigan.
2008-10-07
If you can't lick them
This announcement of the purchase of Biomed Central by Springer is interesting. Who is co-opting whom? Is this acknowledgement of the commercial value of the open access model? Or is it the harbinger of spiraling author fees? Regardless of the motivations or goals, the nature of the editorial boards and contributors to Biomed Central is likely to making Springer tread lightly. Else, alternatives will be generated by the increasingly fluid market of publication venues.
2008-10-06
Snatching defeat from the open (source) jaws of victory?
I have used Endnote for at least a decade as my primary bibliographic tool, long before it was acquired by Thompson Reuters. If the reporting of a lawsuit brought by Thompson Reuters is correct, then as an academic community we need to seriously reconsider our prior recommendations of the use of a product that seems to now be configured precisely against the emerging fluidity of referencing and hyperlinking encouraged by the web from its outset.
One of the widely recognized successes of the Web was indeed in its dissemination of several decades of developments in hyperlinking that allowed, among other uses, different sources of knowledge and information to be hyperlinked. The occasionally wobbly efforts in deploying a Semantic Web that includes some minimalistic formalism of knowledge representation constitute an important and worthy attempt to make such hyperlinking and annotation even more efficient and productive. So, when Thompson starts suing open sourced efforts (using Semantic Web standards) to interoperate with the Endnote bibliographic styles, it is (again if the reports are accurate) creating obstacles to the free flow of information between the richly growing ecosystem of reference and bibliographic applications (web-based or otherwise). This runs counter to all the trends in open source publishing and widely shared document formats.
If indeed, I have misunderstood the nature of the lawsuit then I will readily and publicly retract these comments in this forum. Otherwise, those of us who want our students and colleagues to be able to freely exchange their bibliographic data will consider some alternatives.
2008-10-01
Academia can lead in setting the example for sharing of research data
This article summarizes the benefits of data sharing for research and makes a few common sense recommendations (excerpted below). If our leading academic health centers would adopt these, the yield to all of us (as consumers of research) of our investment in research would grow rapidly.
RECOMMENDATIONS FOR ACADEMIC HEALTH CENTERS TO ENCOURAGE DATA SHARING
- Commit to sharing research data as openly as possible, given privacy constraints. Streamline IRB, technology transfer, and information technology policies and procedures accordingly.
- Recognize data sharing contributions in hiring and promotion decisions, perhaps as a bonus to a publication's impact factor. Use concrete metrics when available.
- Educate trainees and current investigators on responsible data sharing and reuse practices through class work, mentorship, and professional development. Promote a framework for deciding upon appropriate data sharing mechanisms.
- Encourage data sharing practices as part of publication policies. Lobby for explicit and enforceable policies in journal and conference instructions, to both authors and peer reviewers.
- Encourage data sharing plans as part of funding policies. Lobby for appropriate data sharing requirements by funders, and recommend that they assess a proposal's data sharing plan as part of its scientific contribution.
- Fund the costs of data sharing, support for repositories, adoption of sharing infrastructure and metrics, and research into best practices through federal grants and AHC funds.
- Publish experiences in data sharing to facilitate the exchange of best practices.
2008-09-25
Unnecessary thievery
Several years ago, I was working on modeling the hypothalamic-pituitary axis with my colleague Joe Gonzalez-Heydrich. Unsurprisingly, we could not find any primary data in articles ostensibly describing the relationship between various hormones of this axis. So, I found a very nice shareware program called DataThief. DataThief is "a program to extract (reverse engineer) data points from a graph. Typically, you scan a graph from a publication, load it into DataThief, and save the resulting coordinates, so you can use them in calculations or graphs that include your own data." It worked as billed and recently when I was working with my colleague Asher Schacter on predicting outcomes of drug development from pre-clinical data, I remembered how useful DataThief had been and recommended that he use it to extract the primary data from publications for each of the pharmaceuticals he wanted to study. Lo and behold, it worked again!
If only we had a policy in place that required that all primary data be deposited in a public electronic repository or repositories, then this additional, laborious, and time-consuming step would be unnecessary. Bioinformaticians have been very effective in demonstrating the value of sharing primary experimental data (e.g. high throughput data such as gene expression data or gene variant data) but clinical researchers have yet to achieve the same enlightenment. Until then, please make sure your graphs are very accurate in your publications so that others may benefit from your hard work and the taxpayers' investments in your research.
2008-09-10
BLAST this!
David Osterbur often gives extremely well received lectures on the use of public bioinformatics resources for biologists. However, even he is limited in how many audiences he can reach. So, if you know of a biologist who needs some help in the use of BLAST, or the UCSC Genome Browser or even in the search of information regarding herbs and dietary supplements, you will be happy to know that the Countway Library (in collaboration with the MIT Engineering and Sciences Libraries) has made available several instructional videos. Let me know if these are helpful and if you'd like to see more (and about what).
2008-09-08
Out of date but not dated?
This is a great example of how the instant-at-hand-reflexive-cut-and-paste nature of electronic information can bridge the virtual to inflict real harm. Contemplate how clinical out-of-date information can be similarly used to boost the medical malpractice of the incidentalome. Will medical libraries step up to the challenge of keeping the medical profession up to date?
[Thanks to Ben Reis for the pointer]
2008-09-04
The Harvard Catalyst site is live and open
The name of the Harvard University Clinical and Translational Science Center is Catalyst. Several of its resources are publicly available. For example, you can now see the biomedical scholarly output of our university at a glance. You can find people, buildings, phone numbers, directions and parking across the entire University (!) with 18+ participating institutions. You can see the influenza risk across our local geography and recent history. You can explore which clinical trials are supported by the institutions across Catalyst. You can use Webdash to share web pages and publications and their citations with collaborators and colleagues. You can browse and search the available Core facilities (in the hundreds). And if you need analytic help you can reach out to the Catalyst biostatistics program and genetics program, for example. Within a year, we will reveal the data sharing function called SHRINE which allows authorized users to study patient populations (with regulatory oversight) for pharmacovigilance, and various clinical research projects (e.g genome-wide studies of asthma, major depression resistant to standard antidepressants).
This site is the collaborative effort of multiple informatics groups in our community, including HMS Center for Biomedical Informatics, HMS IT, and the IT groups of Partners Healthcare Systems, Beth Israel Deaconess Medical Center, and Children's Hospital. It was an impressive 107 day dash bringing together diverse applications into one package. It's still rough and in progress and I would welcome your comments as would our Research Navigators.
Just some cocktail party conversation for you: Note the relative decline of protein research (relative to other topics) in the past decade at our University. The same indicators (gratifyingly) show the rise of mathematical topics in our life sciences scholarly output. Our most prolific author is Walt Willet (note the alternate ways his name appears each with its own publication history: to be fixed in the next iteration of Medvane). Note that JBC appears to be a popular journal for our authors to publish in.
2008-08-29
Information disclosure makes the case for information altruists
This just-published article in PLoS Genetics by David Craig and Nils Homer reveals how the straightforward use of information technology puts the identify and health risks of individuals within the access of the public if two conditions are fulfilled: 1) Their genome-wide data (e.g. from a SNP chip) was published online and 2) Someone has some DNA from that individual (e.g. life insurance company? Forensic experts?). This is essentially the genomic equivalent of the disclosure mechanisms that Latanya Sweeney highlighted in the case of conventional medical data. As a result of this article, several national research organizations, private and public, are now pulling data down from their websites. This is going to therefore result in at least a temporary setback in genomic data dissemination for research purposes. Which is going to sadden all of us who are working to bring biomedicine forward into the 21st century. In this context, it seems that we will really have to find large cohorts of health information altruists who are willing to share their data with full understanding of the risks, and perhaps full legislative protection against such risks.
2008-08-22
Does electronic publishing diminish the breadth of the scientist's scholarly attention span?
More genetics in high school than medical school?
This highly amusing story about fish that are not what they are said to be, should be a sobering wake-up call to medical schools. Here we read about students using third parties to DNA sequence and then taxonomize samples they procured in restaurants and groceries. In the article we read of high-school students (who are not necessarily interested in careers in science) whose scientific literacy with regard to genetics would put many physicians to shame. Admittedly, these students have privileged access to well-informed thought leaders and yet can we point to equally creative and hands on teaching of genetics (and its commoditization) in our elite medical schools? The gap between the public's knowledge of genetics and that of the "professionals" appears to be continually narrowing even while public expectations of the value of such knowledge continues to grow.
2008-07-28
Human and machine readable attribution
Whenever we post information on any electronic network, there are at least two audiences: human beings (typically viewing the information through a web browser) and automated agents (e.g. web crawlers). Until recently those who wished to inforrm these two audiences of any use restrictions or intellectual property had to do so twice: in machine readable form and in human readable form. One of the problems in having two forms is that with time (or even from the start) they may not represent the same restrictions or openness. That can lead to, at the very least, misunderstandings and annoyance. Fortunately, one of the more useful ways of the Semantic Web is how it allows for a flexible combination of representation that allows both audiences to be served in the same expression. Here is a particularly useful and recent example.
2008-07-22
Collective editing of biological pathways
Following on the successful model of wikipedia a previously centrally managed biological pathways curation activity (e.g. genmapp) has now gone fully community based in the wikipathways project. This is an ambitious project at many levels. The least of the challenges is the technical, how to allow group editing of a connectivity graph? This has been implemented, quite successfully at first glance, by using a Java applet (i.e. called from within the browser). The greatest challenge will be of course a) getting a critical mass of annotators and b) getting collegiality among these collaborators without allow the religious wars that tend to break out over the smallest of disagreements of the appropriate way to represent knowledge. With regard to the former, I note that there already appears to be a community forming around the annotation of apoptosis pathways but when I searched for POMC, nothing was returned although I could find some of the receptors related to that peptide here.
So, it's up to us to make it successful or not. We'll see if the organizers of this resource have found the sweet spot for such a collaborative effort. Here's hoping they have.
2008-07-21
Librarians and translational research: One year of accommodation
Given how central informatics is to the Harvard University CTSA proposal, the directors of the HMS Countway Library of Medicine (who also happen to be co-directors of the HMS Center for Biomedical Informatics) recently decided to help out with a challenging problem: Where to house the CTSA leadership (including Lee Nadler and Steve Freedman) until the University will have prepared their more permanent home next year? We (Alexa McCray and myself) offered to give up our offices on the fifth floor of the Library for one year and relocate ourselves on the fourth floor for that one year. Lee and Steve promised not to get too comfortable in our Library and Daniel Ennis of the administration assured us of the efforts made to create a home elsewhere for our CTSA colleagues.
2008-07-03
Aging and the C-section
This study is a nice example of how we can track secular trends through publicly accreted data. As we have children later in life it may well be that we are running against some biological limits that Obstetric surgery allows us to overcome. As we instrument the healthcare enterprise using informatics technologies, more and more such testable hypotheses are going to be generated. Will we have the governance in place throughout our healthcare systems to test these hypotheses in a timely and responsible manner? Do we have the expertise and tools in place?
2008-06-24
Redefinition of community standards through evidence
This article in the NY Times (free registration required) reports on how Google is planning to use the frequency of search terms in various US communities to show that those previously defined as being outside the community standard are in fact more frequently used than "apple pie". On the one hand, this trend is likely to grow and may likely do so even in medicine (to define the standard of care from the data rather than from the experts interpreting the data). On the other hand, just because the majority follows a particular practice does not make it best, optimal, desirable or even necessarily permissible. It does however, shed considerable light on double standards of various stripes.
2008-06-23
Blogosphere mapped!
This map of the blogosphere reveals both the political lay of the very densely populated land of blogs and the degrees of authoritativeness that each blog appears to hold. Where is the equivalent for the medical bibliome?
2008-06-17
Pubmed central compliance trends
Just heard some very positive news from David Lipman at the NCBI. It does seem that investigators are responding very positively to the new mandate. Just in the last month, author submissions to Pubmed Central (PMC) have increased by at least a factor of five. The author-contributed manuscripts now exceed the journal contributed manuscripts. It also appears that at least 60% of the expected manuscripts are being submitted to PMC. NIH appears to be well on the way to capturing the vast majority of the published output of NIH-funded science which is a wonderful result for the scientific community and the public which it serves.
2008-06-05
Harvard CTSC
2008-05-23
Important leadership for Harvard in open access publishing
Stuart Shieber, a professor of computer science at the Faculty of Arts and Sciences (FAS) has now assumed the leadership of the new Office for Scholarly Communication. This is the natural outgrowth of his leadership of the Provost’s Committee on Scholarly Communication and in making the case for Open Access publishing adoption at the FAS.
Meta-data strikes back?
Ben Adida writes in his blog about a very interesting development in the Yahoo search infrastructure. The bottom-line is that by opening up some of the search results processing through metadata-level (i.e. rdf) processing, Yahoo has enabled a much more personalized user experience. Now, we'll see if the developer community runs with this opportunity.
2008-05-22
A skeptical view of electronic health record benefits
This report from the Congressional Budget Office (CBO) was widely reported in the press today. The press has mainly focused on the report's skepticism about earlier estimates of greater than $40 billion/year savings from broad national electronic health record adoption. This certainly is going to remain a point of controversy but other questions raised by the report merit broader debate. Do EHR's reduce duplicate ordering of tests and reduce adverse events in the outpatient setting? It has been my own intuition that it does, but this CBO report points out some contrary evidence. It seems that these questions constitute a useful research agenda for the medical informatics community which should be further pursued, in many healthcare delivery settings.
2008-05-21
Genetic discrimination is a crime
The Genetic Information Non-Discrimination Act (GINA) was signed into law by President Bush. This is an important first step in moving towards making disclosure of genetic information no more (and no less) concerning than disclosure of medical and family history. All in all a positive step for harnessing the clinical fruit of the genomic revolution.
2008-05-18
Get yourself or your post-doc in the 21st century.
For those of you engaged in genome-scale studies but not completely up to speed in Bioconductor and R. This very short course will be "conducted" by one of the leaders of the Bioconductor project (Vince Carey). Thanks to Vince, this short course is free of charge but you do have to register.
Title:
Statistical computing for genome-scale biology:
An introduction to R and Bioconductor 2.2
When: 27 and 29 May from 1230pm to 3pm.
Countway Medical Library: 4th floor
This course is intended to acquaint biologists and
bioinformaticians with principles and methods of
computing with genome-scale experimental data using
Bioconductor 2.2. Registered students will have access
to media for installing current packages used in the
course. Topics to be covered on the first day include:
high-level introduction to facilities for differential
expression, gene sets, genetics of gene expression,
measurement of CNV; sketch of the R 2.7 language and
analysis environment; Bioconductor containers and
annotation facilities. The second day will be devoted
to case studies in differential expression, genetics
of gene expression, analysis of CNV.
2008-05-01
Rockfeller leads: Creative Commons License Goes Mainstream for Science
As noted in this announcement from Science Commons:
"The [Rockfeller] Press adopted a new copyright policy that returns essential freedoms to authors and extends permissions to the public that are vital to advancing science. This new policy covers its journals, which include the prestigious Journal of Cell Biology, The Journal of Experimental Medicine and The Journal of General Physiology."
See the original announcement for details
2008-04-16
Get arrested and never get lost again.
This comprehensive collection of DNA samples obtained from individuals arrested by an agent of a federal law enforcement agency will have several remarkable consequences. For example, if an information altruist, such as a volunteer for the Personal Genome Project, puts put on the web a substantial fraction of her genome, federal authorities will be able to trivially run a search program to see if any of them match the genomic characteristics of one of the previously arrested individuals. High-throughput genomics finally meets high-throughput forensics.
2008-04-02
Help for Public Access Submissions
Today we launched a series of NIH Public Access Policy pages on the Countway web site. This is the official web site for the university’s guidance on the NIH policy, which goes into effect on April 7, 2008. The site represents a collaboration with the university’s Office of the General Counsel, the university sponsored programs offices, and our very own staff. Special thanks are due to Alexa McCray for her leadership in this matter and to David Hummel, Scott Lapinski, Doug Macfadden, and Halip Saifi, for creating this terrific resource under enormous time pressure. Please take a look at the site (https://www.countway.harvard.edu/publicaccess) when you have a chance.
2008-03-31
What is the evidence?
This recent announcement of the lack of efficacy of a widely prescribed "cholesterol lowering" combination (two drugs) agent should give us pause. Those of us who practice medicine know all too well how much of what we do is art and not science. Despite billions of dollars of research that linked blood biomarkers such as LDL and CRP to heart disease, we now have a well-run trial that seems to show that the lowering of these "bad" biomarkers does not affect thickening of the walls of arteries in a manner previously thought to result in disease. This once again points to the importance of unimpeachable curation of medical evidence and its clear and untrammeled communication to patients and providers alike.
2008-03-27
No sound before its time?
This late breaking story about the recovery of the sound of a French recording, predating Edison's famous recording has relevance to our modern efforts in digital document archiving. Apparently, Edouard-Leon Scott was able to record sound but not in a way that his contemporaries could play back. It makes the point that archives that do not provide for an immediate "read out" can easily be lost to posterity even if they are physically durably accessible. This is the distinction between light and dark archives.
2008-03-23
Notable Book: Inhuman Research, 5.13.08
Every year, we showcase 3-5 notable books. This one, written by Alfred Pasternak, I discovered through the outreach efforts of the Wiesenthal Museum for Tolerance in LA. Dr. Pasternak will be joining us on May 13th for a presentation at 4pm, followed by book signing. He will be accompanied by Ms. Liebe Geft, the director of the Museum of Tolerance, so I am quite sure it should make for a very interesting couple of hours.

2008-03-16
Large-scale extraction of gene-level physiology from the bibliome
Technorati Tags: Informatics, Scholarship
Absence of a plan
Technorati Tags: archives, Collections, Open Access
2008-03-05
HMS Rewind: Harvard Medical School 1997
Technorati Tags: archives, Collections, History, Scholarship
2008-03-03
Do we have enough informaticians?
Technorati Tags: academics, Informatics
Reference your open access articles for your NIH grants starting April 11th.
Technorati Tags: academics, Open Access
2008-02-28
Electronic Medical Records: The Promise That Keeps on Promising
Technorati Tags: healthcare, Informatics
2008-02-19
Multilingual curation of worldwide outbreaks
Technorati Tags: epidemiology, Informatics, Scholarship, Study design, world health
2008-02-18
Overcoming physical space limitations
Technorati Tags: archives, Collections, consumer
2008-02-14
Open Access accounting?
Technorati Tags: economics, Open Access
2008-02-12
The Case for Open Access
Technorati Tags: academics, Open Access
2008-02-11
Accounting for Academics
Technorati Tags: academics, Scholarship
2008-02-10
Archival black hole
2008-02-05
Modern perspective on contagion.
Technorati Tags: Collections, healthcare, History, Scholarship
2008-02-03
Autonomy and guardianship of our own clinical information.
Technorati Tags: consumer, economics, healthcare, Informatics
2008-02-01
Copyright and open access.
Technorati Tags: Open Access, Scholarship
2008-01-27
Interrupted in mid-sentence: Judah Folkman
- Pioneering work on novel materials for anesthesia.
- Surgical treatment of a variety of structural birth defects.
- Ground breaking insights regarding angiogenesis and malignancy.
- New vehicles for the delivery of drugs.
- Novel therapies for glycogen storage disease.
- Early insights into the effects of structure and microenvironments on development and tumorigenesis.
- Champion of the priority of the doctor's obligation to the patient.
- Targeting of non-cancerous diseases for (anti)angiogenic therapy.
- The understanding of how we all live riddled with dormant micromalignancies.
- Recurrent generosity in sharing credit and collaboration.
- Identification of promising antiangiogenic agents.
- Perpetual optimism
Technorati Tags: In memoriam, Noteworthy, Scholarship