Even in the rarefied reaches of the ivory tower, you will find various previously productive scientists with their hands under the table banging on their personal communication devices. In that context, this report on the failure rate of these devices may be of interest. Nonetheless, there does appear to be real functionality that is well suited for scholarly communication and education.
Harvard Medical School's Countway Library of Medicine and the new frontiers in biomedical computing
2008-11-26
2008-11-24
Primary care burdened by primary data
This report out of the Physicians' Foundation shows that doctors are dissatisfied, are going to quit in droves and 94% said the time they devote to non-clinical paperwork in the last three years has increased. So why does it seem that so many are still trying to get into medical school? Is there a medical practice reality chasm?
'Spinning' up the tissues at Harvard
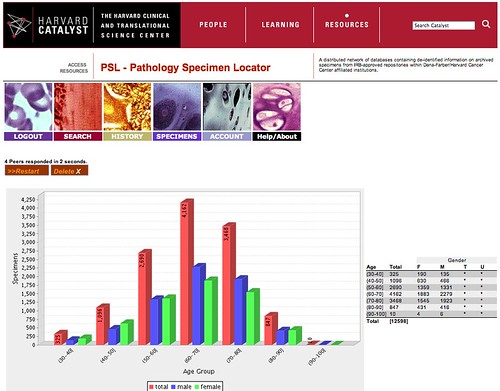
Ever since the late 1990's we have been working on a variety of methods for retrieving data across disparate institutions that are often not even part of the same corporation. In response to an RFA from the National Cancer Institute, called the Shared Pathology Informatics Network, we developed a toolkit that is now in widespread use, specifically to enable genomic and other biological studies on the millions of specimens that are archived across healthcare institutions. As is often the case, we were late in bringing our own tools to use in our own backyard, but that has now happened. The Pathology Specimen Locator (PSL) is now live at our CTSA Catalyst portal. As shown in this screenshot, with authorized credentials, I was able to see that there are over 10,000 lung cancer samples (you can query for any tissue type and disease) across a wide range of ages. It is this sort of IRB-protected, informatics-enabled data liquidity that will accelerate our translational research efforts. Hats off to the entire team but particularly Andy, Frank, John, and Mark.
Transcriptome of a Trio
Hugh Rienhoff will be speaking about an interesting intersection of expression and genetic transmission studies on December 4th.
2008-11-16
Thousands of free books
As per the Lexcyle website "Stanza is a free application for your iPhone and iPod Touch. Use it to download from a vast selection of over 40,000 books and periodicals, and read them right on your phone. It’s a wireless electronic library that stays open 24/7." A quick search for science fiction reveals classics by H. G. Wells and pulp fiction by "Doc" E. E. Smith. All without Digital Rights Management. A real treat.
Images in Development
If an image is worth a thousand words, and a video is 30 frames per second, then these movies of a developing zebrafish must be worth millions. I could not conceive now of a course in embryogenesis that did not include these fascinating and wholistic perspectives on the developing organism. What drives those pulses of development, those three- to four-cell cabals, those intricate dances of the somites? Is it all gradients and cell-cell interactions? Or is there a master control program?
2008-11-12
Matching high-throughput genomics to high-throughput mining of the literature
In this study (alas, for-fee-access) , Dennis Wall demonstrates how to mine the literature (aka the biomedical bibliome) to focus the analyses of noisy genomic modalities which by virtue of measuring thousands of genes have to be aggressively corrected for multiple hypothesis testing [ed. Disclosure: I am a co-author]. By examining gene expression analyses of individuals with autism through the lens of the prior literature on neuro-psychiatric-behavioral disorders, he is able to identify genes significantly differentially expressed in individuals with autism, both known and previously non-implicated genes. This is one of a growing list of publications that are attempting to match the high throughput qualities of genomic measurements with an equally efficient automated "reading" of all the painstakingly obtained biomedical investigational literature. It also suggests that an even more detailed annotation by librarians of the existing literature (analogously to what the National Library of Medicine has done for years for the broad addition of meta data) will be productively leveraged in future investigations. I suppose this is where my colleagues from the Semantic Web have another opportunity to feed the search engines of Google.
2008-11-06
When consent gets in the way
"When consent gets in the way" is the purposefully provocative title of the article (alas, freely open to the public for only 1 month) by Patrick Taylor in which he questions the current dogma about the relationship between consent, privacy, ethical behavior and the public good. Although I was among the early promoters of a strong model of patient personal control of their healthcare data and healthcare decision-making, I found several of Taylor's points compelling. Notably, that "it is questionable whether consent-for-everything will promote privacy and public trust" and "There is more to ethical decision-making than asking whether decisions are made autonomously. Do they take into account virtues, moral values and human narratives with less impoverished conceptions of human freedom? Are the choices good, and do they respect ethical obligations to others?" These points are at the heart of current trends in increasing the "liquidity of patient data" and are certainly central to the business plans of several large companies. How we respond to these assertions as a individuals and a society will be telling. Not only for our research policies and infrastructure but for our conception of healthcare.
2008-11-04
E-science = Librarians 2.0?
In this article (free, but requires registration), in The Scientist, we are given another glimpse into the human components of E-science. I'ts one thing to have a distributed computational network capable of delivering teraflops at the tap of a button, it's another thing to have a biological scientific workforce that knows how to use those cycles at all, let alone effectively. Librarians such as our very own David Osterbur, who are also trained in the biological sciences (and particularly in the use of bioinformatics tools) are a remarkably efficient means to help disseminate a working knowledge of these essential tools to our local communities of biological investigators and beyond.